The three major DESTINA technologies are unique and distinguishable from ALL existing enzymatic methods of nucleic acid analysis.
Art of DGL-Tech™
A game changing technology for detecting nucleic acids and their mutations.
DGL-Tech™ for nucleic acid analysis combines patented SMART-Bases with unique peptide nucleic acid (PNA) capture probes with ‘blank’ positions (DGL-Probes).
The revolutionary DGL-Tech™ works thanks to Watson-Crick base pairing (C-G/A-T) that enables the templating of a dynamic reaction on the DGL-Probe.
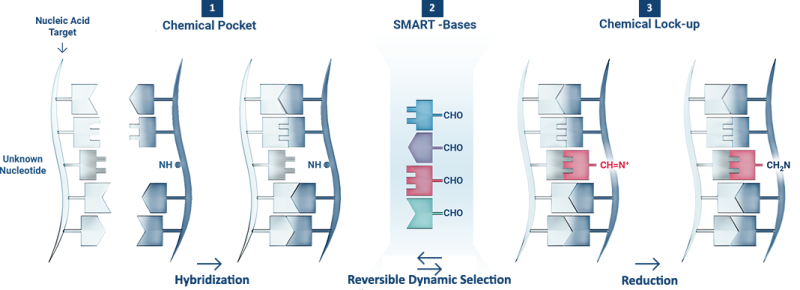
1 The DGL-Probe hybridises with a complementary Nucleic Acid Target, creating a Chemical Pocket containing the Unknown Nucleotide under interrogation.
2 In the presence of four SMART-Bases (i.e. adenine, thymine guanine and cytosine) the Chemical Pocket creates reversible iminium intermediates between the aldehyde chemical group on the SMART-Bases and the amino group on the blank position.
3 The Unknown Nucleotide stabilises the iminium intermediate formed with the complementary SMART-Base. The most stable iminium intermediate is thus reduced (Chemical Lock-up) and analysed.
ChemiRNA™ Tech
Direct detection and absolute quantification of miRNAs from body fluids.
miRNAs are small non-coding RNAs of 19–24 nucleotides in length with a big role in gene regulation. They are present in biological fluids with different expression levels under pathological conditions. miRNAs are rapidly emerging as important biomarkers for clinical diagnosis and prognosis of various human diseases.
Profiling of miRNAs is one of the most active areas of research in the field of liquid biopsy. DESTINA offers a new way to interrogate sequences of miRNAs through its unique ChemiRNA™ Tech.
ChemiRNA™ Tech is based on DESTINA DGL-Tech™ for the direct detection and quantification of miRNAs in body fluids.
- ChemiRNA™ Tech uses a single biotinylated SMART-Base to label the duplex formation with miRNA sequences.
- The technology includes DESTINA proprietary Stabiltech buffer that allows the liberation and stabilisation of RNAs in body fluids, and at the same time removes the need for refrigerated handling up to the analysis.
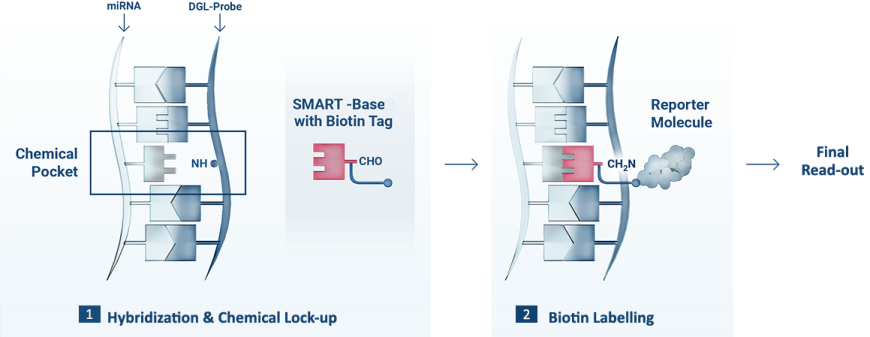
1 The DGL-Probe captures the complementary miRNA sequence, forming the Chemical Pocket. When the target miRNA hybridisation is complete, the biotinylated SMART-Base is covalently attached to the backbone of the DGL-Probe (Chemical Lock-up).
2 The duplex is read out using a Reporter Molecule that recognise the biotin tag.
Compatible ultrasensitive immunoassay platforms
Simplify your RNA analysis with our immunoassay-like platform
SPIN TUBE
DESTINA has developed a novel platform that integrates DGL-Tech™ with a simple colorimetric end-point assay on a porous nylon membrane contained within a micro Spin-Tube.
In the Spin-Tube Device, DGL-Probes are immobilized onto nylon membranes following specific spot patterns. Biotinylated SMART-Bases are used for the labelling of duplexes formed upon the capture of complementary target nucleic acids. Signals are generated by the recognition of the Biotin via streptavidin alkaline phosphatase (Streptavidin-ALP). Incubation with the chromogenic substrate (NBT/BCIP) generates a blue precipitate. Data analyses can be carried out by the naked-eye.

Intellectual Property