The Power Behind PCR-Free Diagnostics
Our hardware-agnostic chemistry transforms standard immunoassay platforms into high-value molecular workstations. By enabling direct, extraction-free detection of miRNAs, isomiRs, and proteins on any readout hardware, DGL-Tech provides the strategic bridge between proteomics and the future of RNA therapeutics.
Strategic Value Pillars
- isomiR Precision & PCR Bias: Absolute precision where PCR fail due to enzymatic bias, ensuring robust data integrity for FDA/EMA submissions.
- Multi-Omic (Proteogenomic) Capability: Combine genomics and proteomics in a single-well reaction using a standard, extraction-free immunoassay procedure.
- Strategic Portfolio Revitalization: Transform generic instrument platforms into high-value molecular workstations by enabling miRNAs detection on immunoassay platforms.
- Universal "Hardware-Agnostic" Architecture: Unlock high-margin molecular assays on any existing platform without CAPEX or hardware modifications.
- Biomarker-Agnostic Scalability: Seamlessly detect miRNAs, isomiRs, piRNAs, and SNPs with a single, standardized, and sequence-selective workflow.
- Bridge to RNA Therapeutics (ASOs): Accelerate drug development with direct sequence-selective recognition and functional screening of Antisense Oligonucleotides.
DGL-Tech: The Core Chemistry
DGL-Tech for nucleic acid analysis combines patented SMART-Bases with unique peptide nucleic acid (PNA) capture probes with ‘blank’ positions (DGL-Probes).
The revolutionary DGL-Tech works thanks to Watson-Crick base pairing (C-G/A-T) that enables the templating of a dynamic reaction on the DGL-Probe.
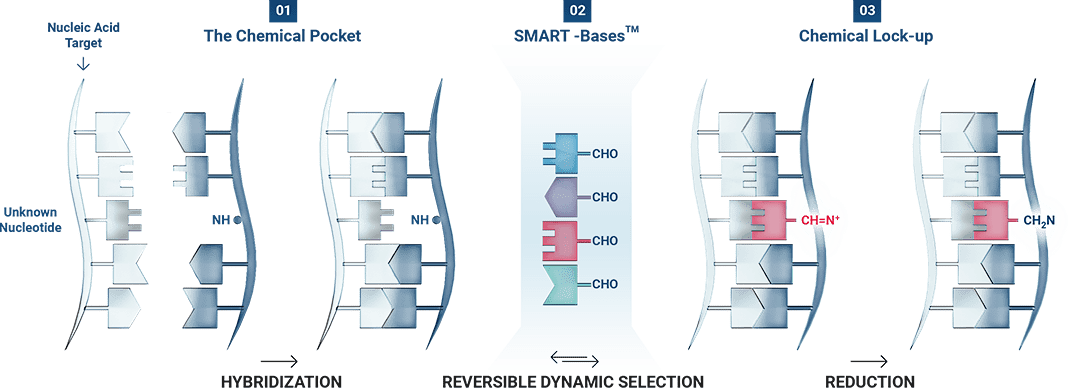
ChemiRNA Tech: The miRNA Application
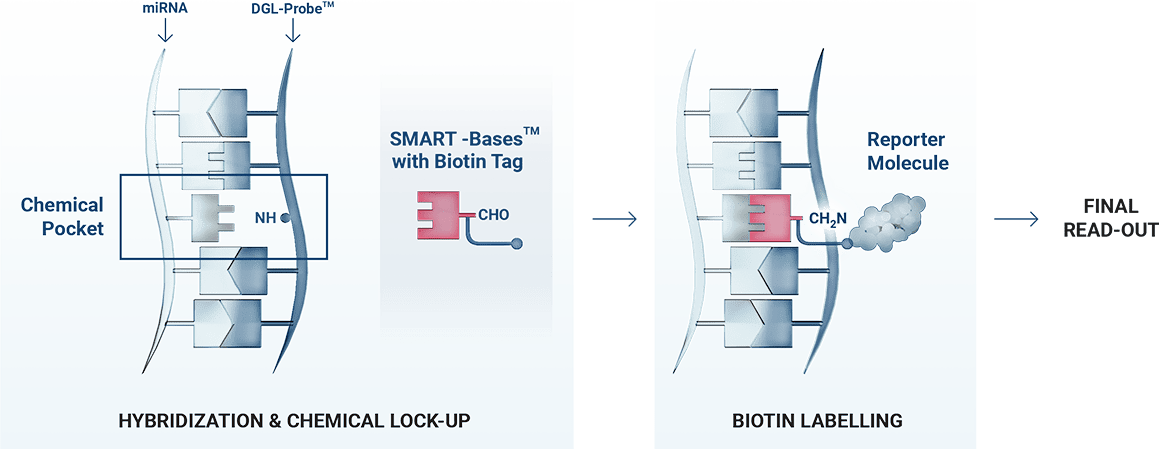
Profiling of miRNAs is one of the most active areas of research in the field of liquid biopsy. DESTINA offers a new way to interrogate sequences of miRNAs through its unique ChemiRNA Tech.
ChemiRNA Tech is based on DESTINA DGL-Tech for the direct detection and quantification of miRNAs in body fluids. It uses a single biotinylated SMART-Base to label the duplex formation with miRNA sequences.
The technology includes DESTINA proprietary Stabiltech buffer that allows the liberation and stabilisation of RNAs in body fluids, and at the same time removes the need for refrigerated handling up to the analysis.
Get Started with DGL-Tech
For Researchers (Pharma/Biotech)
See this technology in action. Test our validated, ready-to-use LiverAce™ kits for DILI safety assessment today.
View LiverAce® Kits →For Partners (CROs/Diagnostics)
License DGL-Tech™ to create your own novel, PCR-Free assays. Gain a competitive advantage in the biomarker market.
View License Opportunities →